My issues with split() in base R
My prior post on tidyverse split-apply-combined ended with me favouring split + map_dfr as a replacement for group_by + do.
In subsequent posts I looked at
the runtime of the split function and
the idioscrasies of the split function and saw the following for the Base R split() function:
- runtime is quadratic in number of splitting variables
- runtime is quadratic in number of groups within each variable
- the splitting variable gets recycled if it’s not as long as the data.frame being split
- NA levels are dropped from the data
Points [1] and [2] mean that split() really slows down as soon as your data grouping becomes moderately complex.
Point [3] means that it’s easy to shoot yourself in the foot and split the data incorrectly.
Point [4] means that split() will just throw away data that happens to have a level value of NA
Why is there no split function in the tidyverse?
According to Hadley on github:
A function that acts rowwise on a data frame doesn’t seem like it should live in purrr.
cleave_by() - a tidyverse-style version ofsplit()
This is an experiment to create a tidyverse-split split() which
avoids the problems listed above. If possible (and with as little work as possible) I want a tidyverse-style split function which
- keeps runtimes linear in the number of variables
- keeps runtimes linear with respect to the number of groups within each variable.
- avoids the recyling of any splitting variable
- keeps NA levels
Note: Since I am simply writing a wrapper around the base split() function, I’m never going to be faster than it for simple grouping examples. This is
because I’m always doing more work upfront which is not necessary for a small number of grouping variables, and only pays off once the number of groups/levels gets
bigger.
#-----------------------------------------------------------------------------
#' tidy version of the base `split()` function
#'
#' @param df data.frame to split
#' @param ... unquoted column names from the 'df' data.frame to specify
#' the splitting groups
#'
#' @return list of data.frames, one for each group
#-----------------------------------------------------------------------------
cleave_by <- function(df, ...) {
stopifnot(inherits(df, "data.frame"))
# use tidyeval to get the names ready for dplyr
grouping <- quos(...)
# Calculate a single number to represent each group
group_index <- df %>%
group_by(!!!grouping) %>%
group_indices()
# do the split by this single group_index variable and return it
split(df, group_index)
}mtcars %>% cleave_by(am, vs)## $`1`
## mpg cyl disp hp drat wt qsec vs am gear carb
## Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2
## Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4
## Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3
## Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3
## Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3
## Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4
## Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4
## Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4
## Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
## AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2
## Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4
## Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2
##
## $`2`
## mpg cyl disp hp drat wt qsec vs am gear carb
## Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1
## Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1
## Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
## Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
## Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4
## Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4
## Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
##
## $`3`
## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4
## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
## Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4
## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6
## Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8
##
## $`4`
## mpg cyl disp hp drat wt qsec vs am gear carb
## Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
## Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
## Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
## Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
## Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
## Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
## Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2cleave_by() keeps NA levels
Creating the groups within cleave_by() uses dplyr::group_by + group_indices.
As both of these functions are aware of (and retain) NA values, using them to create the group labels means that we keep NA levels within a splitting variable.
test_df <- data_frame(a=c(1, 1, 1, 2, 2, 3, NA, NA), b=1:8)## Warning: `data_frame()` is deprecated, use `tibble()`.
## This warning is displayed once per session.test_df %>%
cleave_by(a)## $`1`
## # A tibble: 3 x 2
## a b
## <dbl> <int>
## 1 1 1
## 2 1 2
## 3 1 3
##
## $`2`
## # A tibble: 2 x 2
## a b
## <dbl> <int>
## 1 2 4
## 2 2 5
##
## $`3`
## # A tibble: 1 x 2
## a b
## <dbl> <int>
## 1 3 6
##
## $`4`
## # A tibble: 2 x 2
## a b
## <dbl> <int>
## 1 NA 7
## 2 NA 8cleave_by() avoids factor recycling
split() allows the user to pass in any splitting variable, and in cases where there is a
length mismatch with the number of rows in the original data.frame, it will recycle the values.
With cleave_by(), because we’re using column names of the data.frame to specify the splitting variable, it is
always guaranteed to be the same length as the data.frame itself.
Thus, any recycling of the splitting variable is completely avoided.
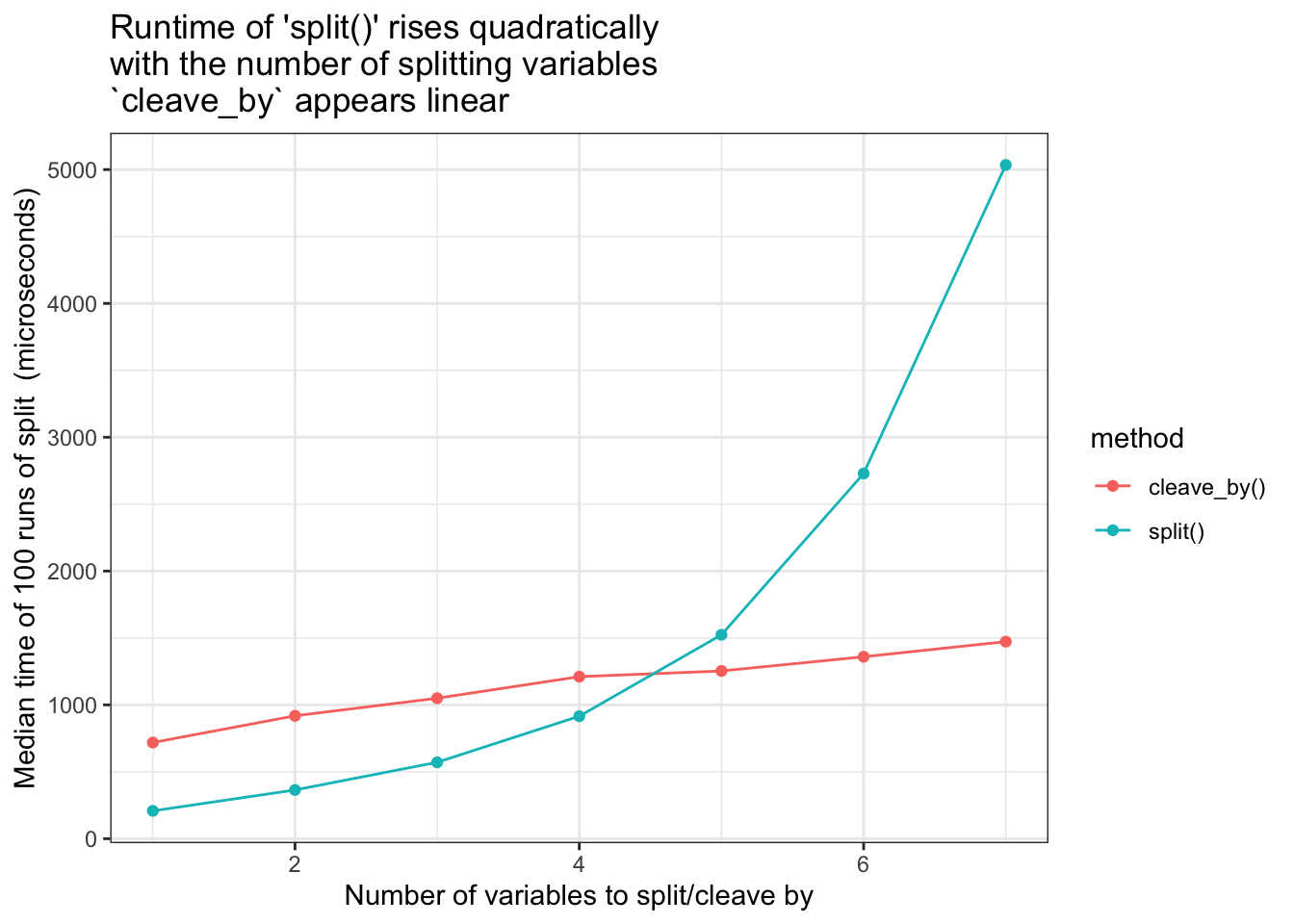
Compare runtimes for increasing number of splitting variables. cleave_by() vs split()
As seen in the graph below, cleave_by() runtimes appear linear with respect to the number of splitting variables, however it does start at a higher baseline time than split().
test_df <- create_test_df(cols=10, rows=10, levels_per_var=2) # See appendix for 'create_test_df'
bench <- microbenchmark(
times = 40,
split(test_df, test_df[, c('a' )]),
split(test_df, test_df[, c('a', 'b' )]),
split(test_df, test_df[, c('a', 'b', 'c' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd', 'e' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd', 'e', 'f' )]),
split(test_df, test_df[, c('a', 'b', 'c', 'd', 'e', 'f', 'g')]),
cleave_by(test_df, a),
cleave_by(test_df, a, b),
cleave_by(test_df, a, b, c),
cleave_by(test_df, a, b, c, d),
cleave_by(test_df, a, b, c, d, e),
cleave_by(test_df, a, b, c, d, e, f),
cleave_by(test_df, a, b, c, d, e, f, g)
)
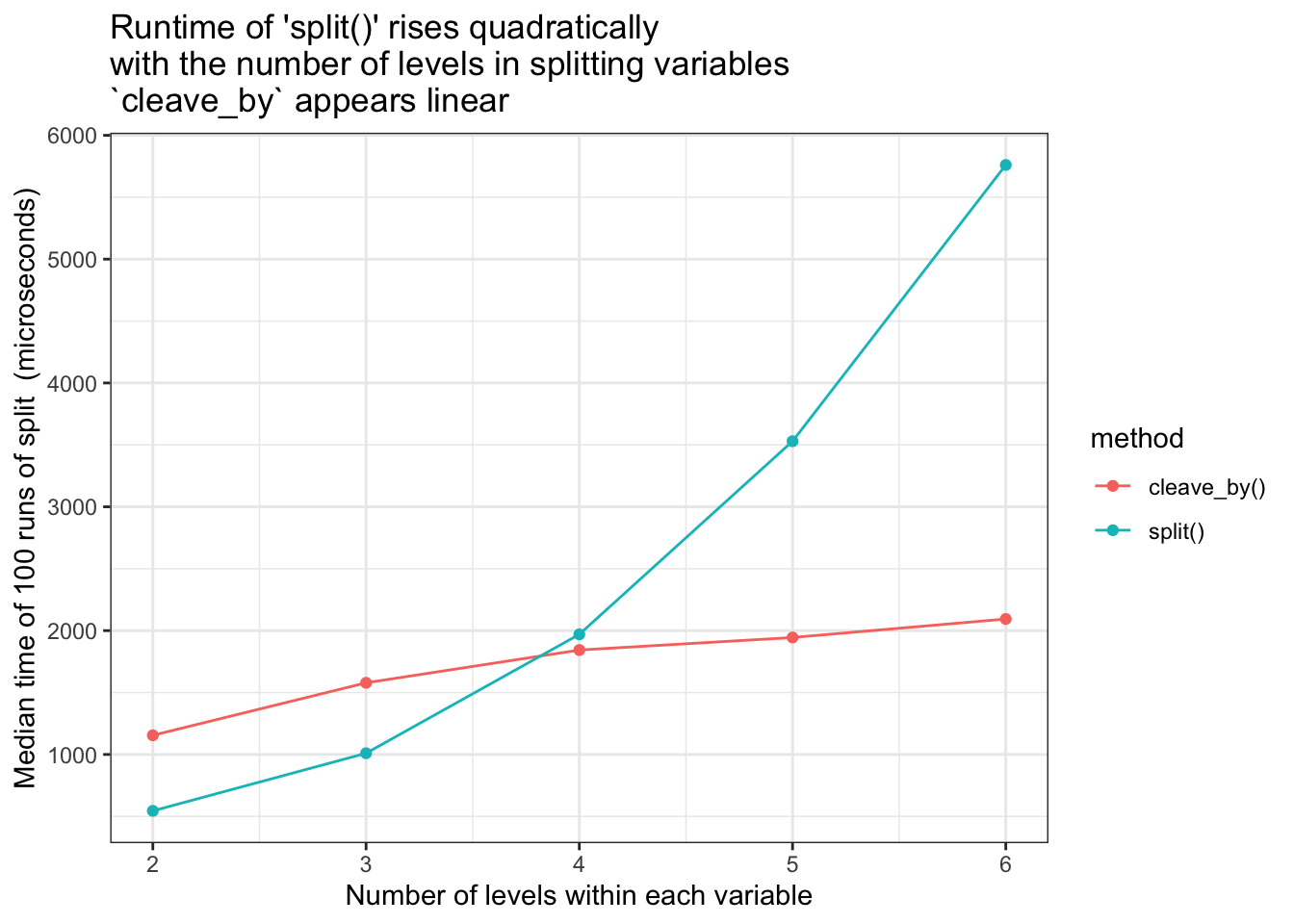
Compare runtimes for increasing number of levels within grouping variables. cleave_by() vs split()
As seen in the graph below, cleave_by() runtimes appear linear with respect to the number of levels within grouping variables, however it does start at a higher baseline time than split()
test_df2 <- create_test_df(cols=4, rows=40, levels_per_var= 2) # See appendix for 'create_test_df'
test_df3 <- create_test_df(cols=4, rows=40, levels_per_var= 3) # See appendix for 'create_test_df'
test_df4 <- create_test_df(cols=4, rows=40, levels_per_var= 4) # See appendix for 'create_test_df'
test_df5 <- create_test_df(cols=4, rows=40, levels_per_var= 5) # See appendix for 'create_test_df'
test_df6 <- create_test_df(cols=4, rows=40, levels_per_var= 6) # See appendix for 'create_test_df'
bench <- microbenchmark(
split(test_df2 , test_df2[, c('a', 'b', 'c')]),
split(test_df3 , test_df3[, c('a', 'b', 'c')]),
split(test_df4 , test_df4[, c('a', 'b', 'c')]),
split(test_df5 , test_df5[, c('a', 'b', 'c')]),
split(test_df6 , test_df6[, c('a', 'b', 'c')]),
cleave_by(test_df2, a, b, c),
cleave_by(test_df3, a, b, c),
cleave_by(test_df4, a, b, c),
cleave_by(test_df5, a, b, c),
cleave_by(test_df6, a, b, c)
)
Conclusions
Does cleave_by() work?
- keeps runtimes linear in the number of variables
- Goal met - the runtime increases linearly not quadratically. But from a higher baseline
- The higher baseline is because of the grouping prep-work which is always done
- This prep-work only pays off when there are more than 4 grouping variables
- Would
Rcpphelp here?
- keeps runtimes linear with respect to the number of groups within each variable.
- Goal met - the runtime increases linearly not quadratically. But from a higher baseline
- The higher baseline is because of the grouping prep-work which is always done
- This prep-work pays off very quickly, and for test dataset
cleave_byhas the advantage oversplit()for 4 or more levels per splitting variable. - Would
Rcpphelp here?
- Avoids the recyling of any splitting variable
- Goal met.
- By only allowing column names to be passed in, if the column name is valid, then the factor length will always be the same as the data.frame length and there will never be value recycling
- Goal met.
- keeps NA levels
dplyr::group_by+group_indicesare both aware of (and keep) NA values. Using them to create the group labels means that NA levels within a splitting variable are kept.
Future
- Would some C++ (via Rcpp) help speed up the baseline case at all?
- Some corner cases to test for:
- What should be done with factor columns with empty levels?
- What about data.frames with zero rows or zero cols?
- Can you
cleave_by()using a list-column as the splitting variable?
Appendix
set.seed(1)
#-----------------------------------------------------------------------------
#' Create a test data set
#'
#' @param cols how many columns?
#' @param rows how many rows?
#' @param levels_per_col how many distinct levels for each variable/column?
#'
#-----------------------------------------------------------------------------
create_test_df <- function(cols, rows, levels_per_var) {
data_source <- letters[seq(levels_per_var)]
create_column <- function(...) {sample(data_source, size = rows, replace = TRUE)}
letters[seq(cols)] %>%
set_names(letters[seq(cols)]) %>%
purrr::map_dfc(create_column)
}