packing-a-data-frame
packing-a-data-frame.RmdsuppressPackageStartupMessages({
library(ggplot2)
library(lobstr)
library(dplyr)
library(purrr)
library(exhibitionist)
library(lofi)
})Introduction
In this vignette, lofi is used to pack each row of the iris data into an integer.
Steps:
- Create a
pack_specfor one row -
pack()/unpack()a single row to test if it works - Use
purrr::map()to apply the packing to every row.
Create a pack spec
The iris dataset gives the measurements in cm of the variables sepal length and width, and petal length and width, respectively, for 50 flowers from each of 3 species of iris. The first rows of the data are shown below:
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species |
|---|---|---|---|---|
| 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 4.7 | 3.2 | 1.3 | 0.2 | setosa |
The pack_spec for the data seen in iris is:
-
Sepal.Lengthis a floating point value with 1 decimal place with a maximum value of 7.9. This could be multiplied by 10, converted to an integer and stored in 7 bits. - Similarly for
Sepal.Width,Petal.LengthandPetal.Width- after multiplying by 10, and treating as an integer, these values could all by stored in 6, 7, and 5 bits respectively. -
Speciesis a choice from 3 options, so in the best case we only need 2 bits to store this information.
The defined pack_spec is stored as a list:
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# Can perfectly pack 'iris' into 27 bits per row.
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
pack_spec <- list(
Sepal.Length = list(type = 'integer', nbits = 7, mult = 10, signed = FALSE),
Sepal.Width = list(type = 'integer', nbits = 6, mult = 10, signed = FALSE),
Petal.Length = list(type = 'integer', nbits = 7, mult = 10, signed = FALSE),
Petal.Width = list(type = 'integer', nbits = 5, mult = 10, signed = FALSE),
Species = list(type = 'choice' , nbits = 2,
options = c('setosa', 'versicolor', 'virginica'))
)Pack/unpack a single row
Now take the first row of iris and pack() it:
So the first row of iris has now been packed into the integer: 54052616.
If this integer is viewed as the 32 bits which make it up, the different lofi data representations can be identified:
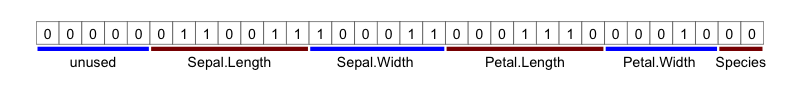
If the integer is now unpack()ed, we get back the original data.
pack/unpack every row
pack/unpack may be mapped over the rows of a data.frame to encode every row as a single integer value.
In the following example, each row of the iris data is encoded as a single 32-bit integer value.
The packed lofi representation of iris is ~12x smaller than the original data.frame.
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# Pack the entire data.frame one row at a time using 'transpose' + 'map'
# `lofi` does not handle factors, so convert 'Species' explicitly to a character
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
iris_packed <- iris %>%
mutate(Species = as.character(Species)) %>%
transpose() %>%
map_int(pack, pack_spec)
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# 'iris' is now encoded as a vector of ints
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
head(iris_packed, 21)
#> [1] 54052616 51873544 49809032 48744328 53020424 57264272 48793356
#> [8] 52987784 46614280 51890052 57231240 50890760 50824964 45581700
#> [15] 61474312 60491664 57263760 54052620 60393612 54101900 57182344
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# Packed representation is smaller by a factor of 10
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
as.numeric(lobstr::obj_size(iris) / lobstr::obj_size(iris_packed))
#> [1] 11.8642
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
# And can unpack the integers into the original data.frame representation
#~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
iris_packed %>%
map(unpack, pack_spec) %>%
bind_rows() %>%
head()
#> # A tibble: 6 x 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <dbl> <dbl> <dbl> <dbl> <chr>
#> 1 5.1 3.5 1.4 0.2 setosa
#> 2 4.9 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
#> 4 4.6 3.1 1.5 0.2 setosa
#> 5 5 3.6 1.4 0.2 setosa
#> 6 5.4 3.9 1.7 0.4 setosa